Benchmarking large-scale integration#
Here we walkthrough applying the integration benchmarking metrics on a dataset of non-small cell lung cancer from the following paper:
Salcher, S., Sturm, G., Horvath, L., Untergasser, G., Kuempers, C., Fotakis, G., … & Trajanoski, Z. (2022). High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell.
This dataset contains ~900,000 cells, so here we demonstrate the scalbility of the metrics here on large data.
import numpy as np
import scanpy as sc
from scvi.data import cellxgene
from scib_metrics.benchmark import Benchmarker, BioConservation
%matplotlib inline
Load and preprocess data#
url = "https://cellxgene.cziscience.com/e/232f6a5a-a04c-4758-a6e8-88ab2e3a6e69.cxg/"
adata = cellxgene(url, filename="luca.h5ad", save_path="data/")
adata
AnnData object with n_obs × n_vars = 892296 × 17811
obs: 'sample', 'uicc_stage', 'ever_smoker', 'age', 'donor_id', 'origin', 'dataset', 'ann_fine', 'cell_type_predicted', 'doublet_status', 'leiden', 'n_genes_by_counts', 'total_counts', 'total_counts_mito', 'pct_counts_mito', 'ann_coarse', 'cell_type_tumor', 'tumor_stage', 'EGFR_mutation', 'TP53_mutation', 'ALK_mutation', 'BRAF_mutation', 'ERBB2_mutation', 'KRAS_mutation', 'ROS_mutation', 'origin_fine', 'study', 'platform', 'cell_type_major', 'suspension_type', 'assay_ontology_term_id', 'cell_type_ontology_term_id', 'development_stage_ontology_term_id', 'disease_ontology_term_id', 'self_reported_ethnicity_ontology_term_id', 'is_primary_data', 'organism_ontology_term_id', 'sex_ontology_term_id', 'tissue_ontology_term_id', 'cell_type', 'assay', 'disease', 'organism', 'sex', 'tissue', 'self_reported_ethnicity', 'development_stage'
var: 'is_highly_variable', 'mito', 'n_cells_by_counts', 'mean_counts', 'pct_dropout_by_counts', 'total_counts', 'feature_is_filtered', 'feature_name', 'feature_reference', 'feature_biotype'
uns: '_scvi', 'ann_fine_colors', 'batch_condition', 'cell_type_major_colors', 'cell_type_ontology_term_id_colors', 'cell_type_predicted_colors', 'dataset_colors', 'default_embedding', 'hvg', 'leiden_colors', 'neighbors', 'origin_colors', 'platform_colors', 'schema_version', 'title', 'umap'
obsm: 'X_scANVI', 'X_scVI', 'X_umap'
layers: 'count', 'counts_length_scaled'
obsp: 'connectivities', 'distances'
sc.pp.subsample(adata, n_obs=500_000)
# Number of unique cell types
adata.obs["cell_type"].nunique()
33
adata.var["highly_variable"] = np.asarray(adata.var["is_highly_variable"].astype(bool))
sc.tl.pca(adata)
“Run” methods#
The authors used scVI and scANVI in their manuscript, and these embeddings are already stored in the AnnData object. We can use these to run the metrics.
scVI#
adata.obsm["scVI"] = adata.obsm["X_scVI"]
scANVI#
adata.obsm["scANVI"] = adata.obsm["X_scANVI"]
Perform the benchmark#
Here we use a custom nearest neighbor function to speed up the computation of the metrics. This is not necessary, but can be useful for large datasets.
In particular we use faiss, which can be accelerated with a GPU.
This can be installed as
conda install -c conda-forge faiss-gpu
When using approximate nearest neighbors, an issue can arise where each cell does not get a unique set of K neighbors. This issue happens with faiss hnsw below, so we use the brute force method instead, which is still faster than pynndescent approximate nearest neighbors on CPU.
import faiss
from scib_metrics.nearest_neighbors import NeighborsResults
def faiss_hnsw_nn(X: np.ndarray, k: int):
"""Gpu HNSW nearest neighbor search using faiss.
See https://github.com/nmslib/hnswlib/blob/master/ALGO_PARAMS.md
for index param details.
"""
X = np.ascontiguousarray(X, dtype=np.float32)
res = faiss.StandardGpuResources()
M = 32
index = faiss.IndexHNSWFlat(X.shape[1], M, faiss.METRIC_L2)
gpu_index = faiss.index_cpu_to_gpu(res, 0, index)
gpu_index.add(X)
distances, indices = gpu_index.search(X, k)
del index
del gpu_index
# distances are squared
return NeighborsResults(indices=indices, distances=np.sqrt(distances))
def faiss_brute_force_nn(X: np.ndarray, k: int):
"""Gpu brute force nearest neighbor search using faiss."""
X = np.ascontiguousarray(X, dtype=np.float32)
res = faiss.StandardGpuResources()
index = faiss.IndexFlatL2(X.shape[1])
gpu_index = faiss.index_cpu_to_gpu(res, 0, index)
gpu_index.add(X)
distances, indices = gpu_index.search(X, k)
del index
del gpu_index
# distances are squared
return NeighborsResults(indices=indices, distances=np.sqrt(distances))
import time
adata.obsm["Unintegrated"] = adata.obsm["X_pca"]
biocons = BioConservation(isolated_labels=False)
start = time.time()
bm = Benchmarker(
adata,
batch_key="sample",
label_key="cell_type",
embedding_obsm_keys=["Unintegrated", "scANVI", "scVI"],
pre_integrated_embedding_obsm_key="X_pca",
bio_conservation_metrics=biocons,
n_jobs=-1,
)
bm.prepare(neighbor_computer=faiss_brute_force_nn)
bm.benchmark()
end = time.time()
print(f"Time: {int((end - start) / 60)} min {int((end - start) % 60)} sec")
Time: 20 min 45 sec
Visualize the results#
bm.plot_results_table()
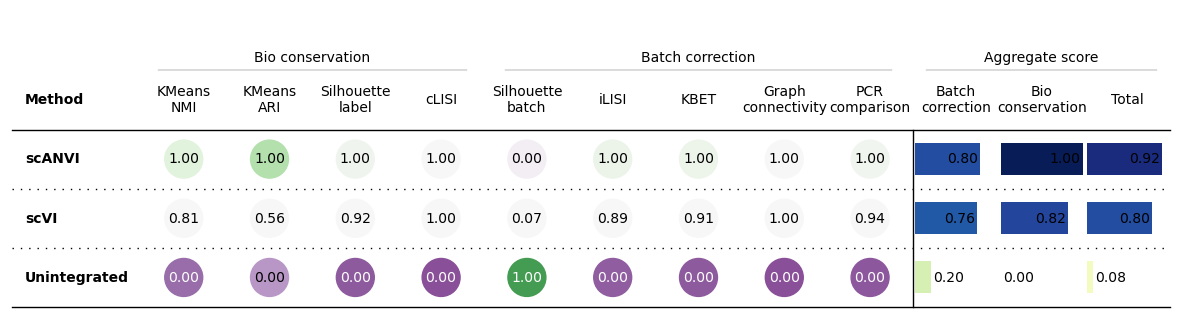
<plottable.table.Table at 0x7f2642db39a0>
bm.plot_results_table(min_max_scale=False)
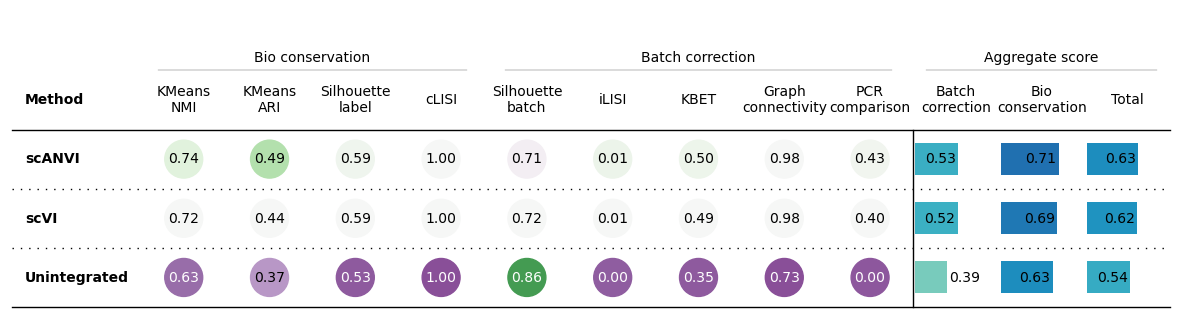
<plottable.table.Table at 0x7f2714881870>
We can also access the underlying dataframes to print the results ourselves.
from rich import print
df = bm.get_results(min_max_scale=False)
print(df)
KMeans NMI KMeans ARI Silhouette label \ Embedding Unintegrated 0.630967 0.372678 0.531423 scANVI 0.744273 0.485717 0.594908 scVI 0.722253 0.435955 0.590104 Metric Type Bio conservation Bio conservation Bio conservation cLISI Silhouette batch iLISI \ Embedding Unintegrated 0.999528 0.861078 0.002044 scANVI 1.0 0.710921 0.009262 scVI 1.0 0.721547 0.008491 Metric Type Bio conservation Batch correction Batch correction KBET Graph connectivity PCR comparison \ Embedding Unintegrated 0.350416 0.733284 0.0 scANVI 0.504855 0.981208 0.428348 scVI 0.490899 0.980226 0.403148 Metric Type Batch correction Batch correction Batch correction Batch correction Bio conservation Total Embedding Unintegrated 0.389365 0.633649 0.535935 scANVI 0.526919 0.706225 0.634502 scVI 0.520862 0.687078 0.620592 Metric Type Aggregate score Aggregate score Aggregate score